
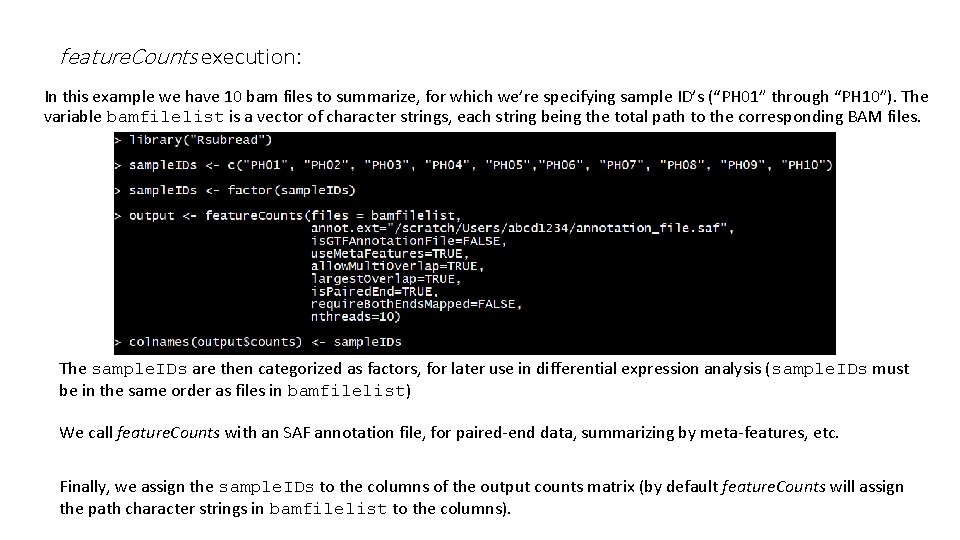
#BAM FILE FORMAT COLUMNS PLUS#
Similarly, the final blockStart position plus theįinal blockSize value must equal chromEnd. In BED files with block definitions, the first blockStart value must be 0, so that the firstīlock begins at chromStart. Items in this list should correspond to blockCount. All of theīlockStart positions should be calculated relative to chromStart.
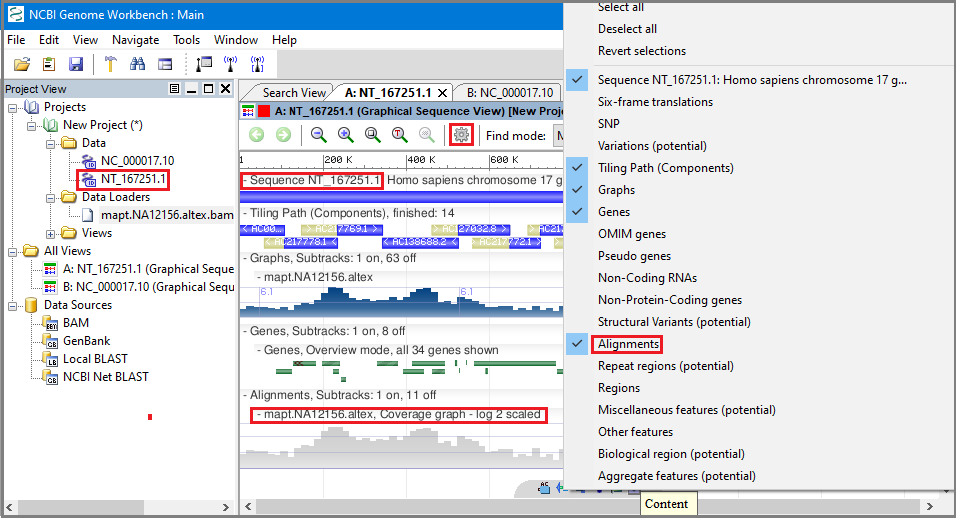
ItemRgb attribute is set to "On", this RBG value will determine the displayĬolor of the data contained in this BED line.
#BAM FILE FORMAT COLUMNS FULL#
The BED line in the Genome Browser window when the track is open to full display mode or directly name - Defines the name of the BED line.The 9 additional optional BED fields are: For example, use chromStart=0, chromEnd=0 to represent an insertion before the
#BAM FILE FORMAT COLUMNS SOFTWARE#
The first 100 bases of chromosome 1 are defined as chrom=1, chromStart=0, chromEnd=100,Īnd span the bases numbered 0-99 in our software (not 0-100), but will represent theĬhromStart and chromEnd can be identical, creating a feature of length 0, commonly The number in position format will be represented. TheĬhromEnd base is not included in the display of the feature, however,
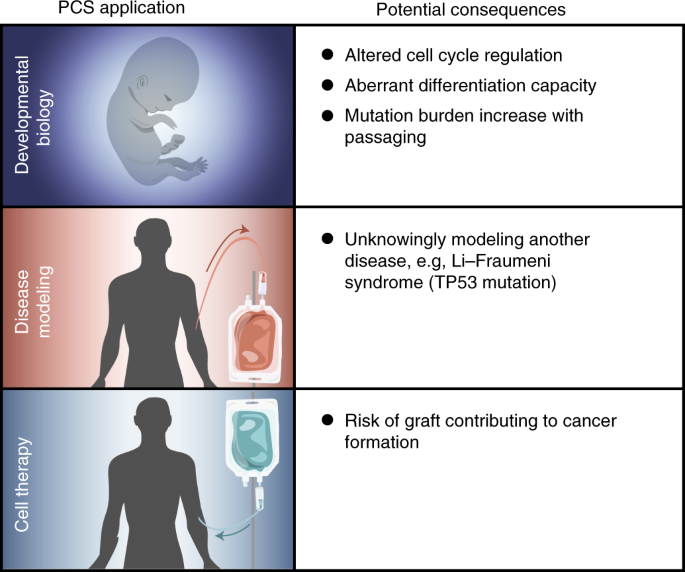
Header lines are not permissible in downstream utilities such as bedToBigBed, which convert lines of Which begin with the word "browser" or "track" to assist the browser in theĭisplay and interpretation of the lines of BED data following the headers. Please note that only in custom tracks can the first lines of the file consist of header lines, Only some variations of BED types, such asīedDetail, require a tab character delimitation for Some circumstances, if the field content is to be empty. The order of the optional fields is binding: lower-numbered fields must alwaysīe populated if higher-numbered fields are used.īED information should not be mixed as explained above (BED3 should not be mixed with BED4), ratherĪdditional column information must be filled for consistency, for example with a "." in The number of fields per line must be consistent throughout any single set of data in anĪnnotation track. BED lines have three required fields and nine additional optionalįields. Return to FAQ Table of Contents BED formatīED (Browser Extensible Data) format provides a flexible way to define the data lines that areĭisplayed in an annotation track.


 0 kommentar(er)
0 kommentar(er)
